DB Schema
The VitaPad schema is best understood if you look at it in two parts: the biological subschema and the graphical subschema. In fact, the design goal here was to separate biological information from graphical information. Loose coupling at this level allows new classes of data, unknown to the original configuration to be added easily to the database. It makes the entire application more flexible and extensible
The Biological Subschema
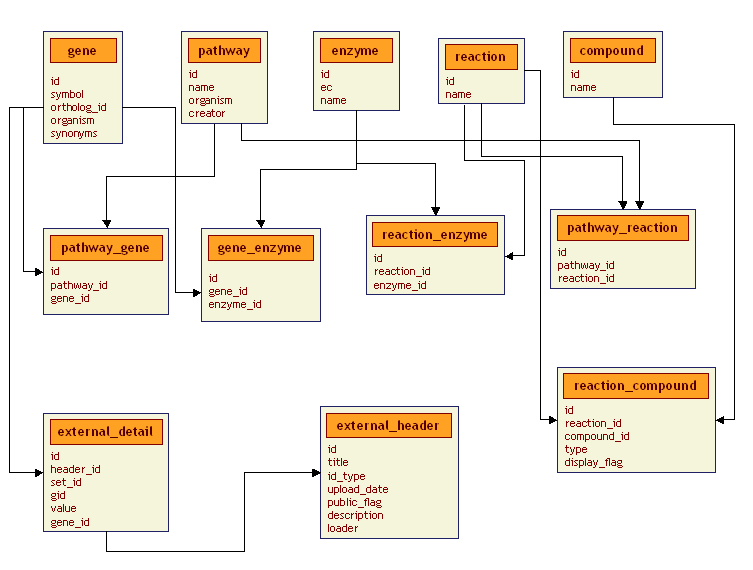
This subschema deals with the scientific details of the pathways we are looking at, things like compounds and genes. It is fairly standard in its construction and its not difficult to conceive of how additional tables could be added to reflect other classes of scientific data. Let's look at the tables:
-
gene table:
- id - The unique, machine-generated identifier of the gene
-
symbol - The symbol assigned (by Affymetrix, LocusLink or whoever) to the gene
-
ortholog-id - The ortholog id of the gene
-
organism - The organism we are looking at
-
synonyms - Optional other names for the gene
-
pathway table:
- id - The unique, machine-generated identifier of the pathway
-
name - The official name of the pathway
-
organism - The organism we are dealing with
-
creator - Who or what created this pathway diagram.
-
enzyme table:
- id - The unique, machine-generated identifier of the enzyme
-
ec - The official E.C. number of the enzyme
-
name - The full name of the enzyme
-
reaction table:
- id - The unique, machine-generated identifier of the reaction
-
name - The name of the reaction
-
compound table:
- id - The unique, machine-generated identifier of the compound
-
name - The name of the compound
-
pathway_gene table:
- id - A unique, machine-generated identifier for indexing
-
pathway_id - A foreign key referencing the pathway table
-
gene_id - A foreign key referencing the gene table
-
gene_enzyme table:
- id - A unique, machine-generated identifier for indexing
-
gene_id - A foreign key referencing the gene table
-
enzyme_id - A foreign key referencing the enzyme table
-
reaction_enzyme table:
- id - A unique, machine-generated identifier for indexing
-
reaction_id - A foreign key referencing the reaction table
-
enzyme_id - A foreign key referencing the enzyme table
-
pathway_reaction table:
- id - A unique, machine-generated identifier for indexing
-
pathway_id - A foreign key referencing the pathway table
-
reaction_id - A foreign key referencing the reaction table
-
reaction_compound table:
- id - A unique, machine-generated identifier for indexing
-
reaction_id - A foreign key referencing the reaction table
-
compound_id - A foreign key referencing the compound table
-
type - Product ('P') or Substrate ('S').
-
display_flag - Make this compound visible on the graph or not
-
external_header table:
- id - A unique, machine-generated identifier for the experiment
-
title - The name of the experiment
-
id_type - What types of id's will be used (Affy, Unigene, etc.)
-
upload_date - When was this experiment submitted to the database?
-
public_flag - Is this public or private data?
-
description - A brief description of the experiment
-
loader - Who loaded the experiment?
-
external_detail table:
- id - A unique, machine-generated identifier for a data point in an experiment
-
header_id - The experiment this data point belongs to. (Foreign key referencing the external_header table)
-
set_id - The replicate number of this data point
-
gid - An official id of the gene (from Affy, Unigene or whatever)
-
value - The value of this data point
-
gene_id - Foreign key referencing the gene table
The Graphical Subschema
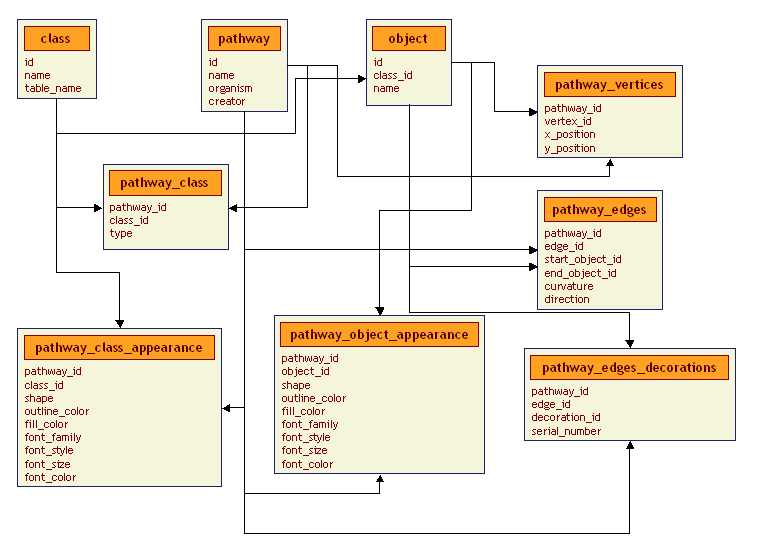
The subscheam dealing with pathway rendering is more complex. It is based upon the EAV/CR (Entity-Attribute-Value with Classes and Relationships) schema that was developed by Prakash Nadkarni, Luis Marenco and others at YCMI. See herefor more information. This design allows for customization of visual components at three levels: global, pathway and individual element.
-
class table: (This is a metadata table. We declare a data class and state which table its information is stored in)
- id - A unique, machine-generated identifier
-
name - The name of the class
-
table_name - The name of the table where data on instances of this class are stored in the biological subschema
-
pathway table: (Duplicated from the biological subschema above)
-
object table: (A central dictionary of rendered elements. All instances of classes have an entry here)
- id - A unique, machine-generated identifier
-
class_id - The class this is an instance of. A foreign key referencing classes
-
name - The name of this object
-
pathway_class table: (This table controls the default drawing type (vertex, edge, decoration) for a class in a particular pathway. A '_DEFAULT' pathway is used to store defaults for all pathways. e.g. The default representation of a gene in a metabolic pathway is as a decoration, but for a signalling pathway it might be preferable as a vertex)
- pathway_id - A foreign key referencing pathway
-
class_id - A foreign key referencing class
-
type - The graphical type (vertex, edge or decoration)
-
pathway_class_appearance table: (This table governs how a certain class is rendered in a pathway. A '_DEFAULT' pathway is used to store defaults for all pathways.)
- pathway_id - A foreign key referenceing pathway
-
class_id - A foreign key referencing class
-
shape - The shape to be used (Rectangle, Ellipse and Rounded Rectangle are currently supported)
-
outline_color - In hex string format #000000-#FFFFFF
-
fill_color - In hex string format #000000-#FFFFFF
-
font_family - The base name of the font (system-dependent)
-
font-style - (0-plain, 1-bold, 2-italic)
-
font_size - In points
-
font_color - In hex string format #000000-#FFFFFF
-
pathway_object_appearance table: (This table governs how an individual object is rendered in a pathway.
- pathway_id - A foreign key referenceing pathway
-
object_id - A foreign key referencing class
-
shape - The shape to be used (Rectangle, Ellipse and Rounded Rectangle are currently supported)
-
outline_color - In hex string format #000000-#FFFFFF
-
fill_color - In hex string format #000000-#FFFFFF
-
font_family - The base name of the font (system-dependent)
-
font-style - (0-plain, 1-bold, 2-italic)
-
font_size - In points
-
font_color - In hex string format #000000-#FFFFFF
-
pathway_vertices table: (Information specific to rendering vertices)
- pathway_id - A foreign key referencing pathway
-
vertex_id - A foreign key referencing object
-
x_position - The x coordinate of the vertex
-
y_position - The y coordinate of the vertex
-
pathway_edges table: (Information specific to rendering edges)
- pathway_id - A foreign key referencing pathway
-
edge_id - A foreign key referencing object
-
start_object_id - The source of the edge
-
end_object_id - The sink of the edge
-
curvature - The curvature of the edge line
-
direction - The direction of the edge line. (F-Forward, R-Reverse, B-Both ways, N-Neither way)
-
pathway_edges_decorations table: (Information specific to rendering decorations)
- pathway_id - A foreign key referencing pathway
-
edge_id - A foreign key referencing object
-
decoration_id - A foreign key referencing object
-
serial_number - The index of this decoration along the edge. Used to determine its relative location to the edge


